Spotting and diagnosing cancer is a complex and difficult process even for the dedicated medical professionals who do it for a living. A new tool from Google researchers could improve the process by providing what amounts to reverse image search for suspicious or known cancerous cells. But it’s more than a simple matching algorithm.
Part of the diagnosis process is often examining tissue samples under a microscope and looking for certain telltale signals or shapes that may indicate one or another form of cancer. This can be a long and arduous process because every cancer and every body is different, and the person inspecting the data must not only look at the patient’s cells but also compare them to known cancerous tissues from a database or even a printed book of samples.
As has been amply demonstrated for years now, matching similar images to one another is a job well-suited to machine learning agents. It’s what powers things like Google’s reverse image search, where you put in one picture and it finds ones that are visually similar. But this technique has also been used to automate processes in medicine, where a computer system can highlight areas of an X-ray or MRI that have patterns or features it has been trained to recognize.
That’s all well and good, but the complexity of cancer pathology rules out simple pattern recognition between two samples. One may be from the pancreas, another from the lung, for example, meaning the two situations might be completely different despite being visually similar. And an experienced doctor’s “intuition” is not to be replaced, nor would the doctor suffer it to be replaced.
Aware of both the opportunities and limitations here, Google’s research team built SMILY (Similar Medical Images Like Yours), which is a sort of heavily augmented reverse image search built specifically for tissue inspection and cancer diagnosis.
A user puts into the system a new sample from a patient — a huge, high-resolution image of a slide on which a dyed section of tissue is laid out. (This method is standardized and has been for a long time — otherwise how could you compare any two?)
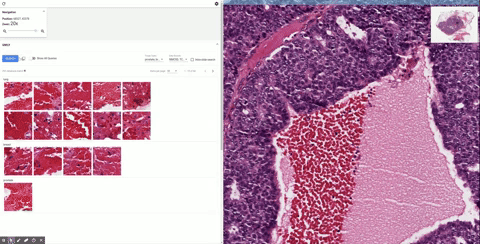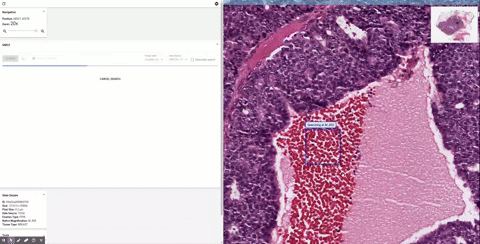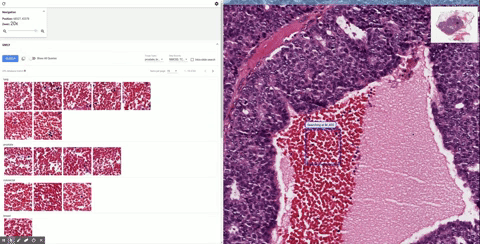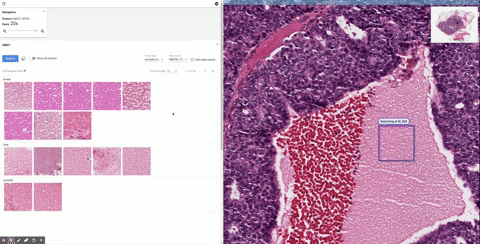
Once it’s in the tool, the doctor can inspect it as they would normally, zooming in and panning around. When they see a section that piques their interest, they can draw a box around it and SMILY will perform its image-matching magic, comparing what’s inside the box to the entire corpus of the Cancer Genome Atlas, a huge database of tagged and anonymized samples.
Similar-looking regions pop up in the sidebar, and the user can easily peruse them. That’s useful enough right there. But as the researchers found out while they were building SMILY, what doctors really needed was to be able to get far more granular in what they were looking for. Overall visual similarity isn’t the only thing that matters; specific features within the square may be what the user is looking for, or certain proportions or types of cells.
As the researchers write:
Users needed the ability to guide and refine the search results on a case-by-case basis in order to actually find what they were looking for…This need for iterative search refinement was rooted in how doctors often perform “iterative diagnosis”—by generating hypotheses, collecting data to test these hypotheses, exploring alternative hypotheses, and revisiting or retesting previous hypotheses in an iterative fashion. It became clear that, for SMILY to meet real user needs, it would need to support a different approach to user interaction.
To this end the team added extra tools that let the user specify much more closely what they are interested in, and therefore what type of results the system should return.
First, a user can select a single shape within the area they are concerned with, and the system will focus only on that, ignoring other features that may only be distractions.
Second, the user can select from among the search results one that seems promising and the system will return more like it, less closely tied to the original query. This lets the user go down a sort of rabbit hole of cell features and types, doing that “iterative” process the researchers mentioned above.
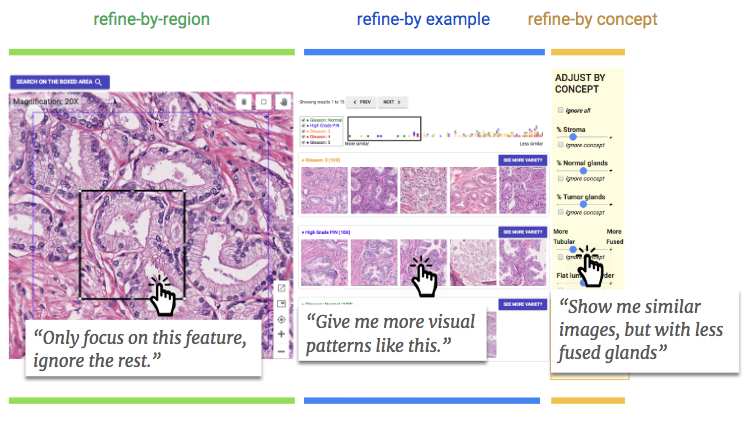
And third, the system was trained to understand when certain features are present in the search result, such as fused glands, tumor precursors and so on. These can be included or excluded in the search — so if someone is sure it’s not related to this or that feature, they can just sweep all those examples off the table.
In a study of pathologists given the tool to use, the results were promising. The doctors appeared to adopt the tool quickly, not only using its official capabilities but doing things like reshaping the query box to test the results or see if their intuition on a feature being common or troubling was right. “The tools were preferred over a traditional interface, without a loss in diagnostic accuracy,” the researchers write in their paper.
It’s a good start, but clearly still only an experiment. The processes used for diagnosis are carefully guarded and vetted; you can’t just bring in a random new tool and change up the whole thing when people’s lives are on the line. Rather, this is merely a bright start for “future human-ML collaborative systems for expert decision-making,” which may at some point be put into service at hospitals and research centers.
You can read the two papers describing SMILY and the doctor-focused refinements to SMILY here; they were originally presented at CHI 2019 in Glasgow earlier this year.